Enhancing Patient Diagnosis with Graph-based Retrieval-Augmented Generation
📚 View all posts in the Graph-based Healthcare Series
Graph-based Healthcare Series — 2
This is the second post in an ongoing series on graph-based healthcare tools. Stay tuned for upcoming entries on clinical modeling, decision support systems, and graph-powered AI assistants.
In our previous post, we explored how the Integrated Management of Neonatal and Childhood Illness (IMNCI) guidelines were transformed from static, text-heavy documents into an interactive graph model. This structure enabled more intuitive navigation of clinical logic, laying the groundwork for advanced applications in AI-assisted patient diagnosis.
In this follow-up, we demonstrate how that graph model serves as the foundation for a graph-based retrieval-augmented generation (graph RAG) system. By combining the structured clinical knowledge encoded in Neo4j with the generative capabilities of large language models (LLMs), we create a framework that supports transparent, context-aware patient diagnosis at the point of care.
From Graph Data Modeling to Graph-based Retrieval Systems
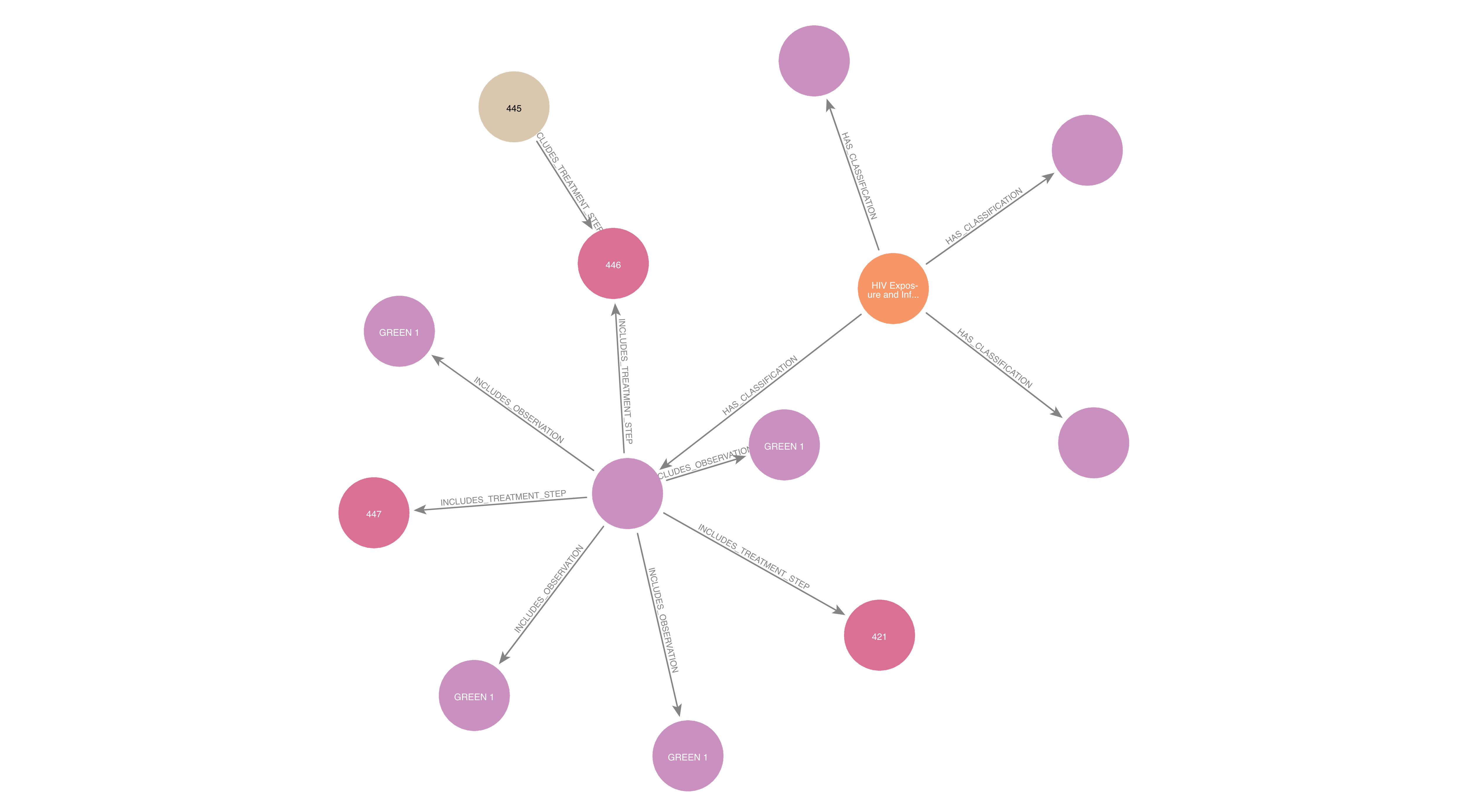
In our prior work modeling the IMNCI guidelines, we developed a comprehensive graph data model in Neo4j that captures the logical structure of pediatric clinical protocols. This model represents key elements of the guidelines as graph nodes—such as Conditions, Classifications, Observations, and Treatment Steps—and encodes their relationships using typed edges that reflect clinical reasoning pathways.
For example:
(Condition) -[:HAS_CLASSIFICATION]-> (Classification)(Classification) -[:INCLUDES_OBSERVATION]-> (Observation)(Classification) -[:INCLUDES_TREATMENT_STEP]-> (Treatment Step)
Example subgraph show the relationships between medical condition, classifications, observations, and treatment steps. Each node type is connected by edges that reflect the logical structure of the guidelines.
These relationships are not merely descriptive—they reflect conditional logic embedded in the original guidelines. In other words, the graph encodes sequences like: “If these symptoms are present, then the medical condition is classified as X, and the recommended treatment is Y.”
With this structure in place, the next challenge is to operationalize the graph for use in clinical decision support tools. This means developing a way to:
- Dynamically query the graph based on patient data (e.g., symptoms, age group, test results, etc.)
- Retrieve clinically relevant subgraphs/reasoning chains that are targeted for specific patient cases
- Use those results to inform and ground responses from LLMs
In the following sections, we describe how we transitioned from a static graph representation to a live, queryable retrieval system that can support complex diagnostic reasoning. This system forms the foundation for our graph RAG pipeline.
Why Graph-based Retrieval for Clinical Decision Support?
Traditional information retrieval methods—such as keyword search or vector similarity—struggle in clinical settings where decisions depend on contextual, conditional, and often multi-step reasoning. Clinical guidelines like IMNCI are rich in logical dependencies: the presence or absence of a symptom may lead to different classifications, which in turn dictate specific treatment paths. Flat text or embedding-based approaches often miss these structured relationships.
Graph-based retrieval provides a more principled alternative by aligning closely with how clinical logic is organized. Using a graph database like Neo4j, we can:
- Traverse complex clinical logic chains: Starting from observed symptoms, we can follow relationships to relevant classifications and their corresponding treatments—capturing multi-hop reasoning paths in a structured, interpretable way.
- Apply logical operators natively: Many IMNCI rules depend on combinations of
conditions using
AND,OR, orIF. These can be represented and queried directly in the graph viaLogicGroupnodes, ensuring high fidelity to the original guideline logic. - Filter and personalize queries: Graph queries can be dynamically adjusted based on patient-specific attributes such as age group or test availability, allowing fine-grained control over retrieved information.
Importantly, graph-based retrieval complements large language models. While LLMs excel at synthesizing and communicating information, graphs ensure that the underlying content is grounded in verified, structured medical knowledge. This hybrid approach—retrieval-augmented generation using a clinical knowledge graph—helps produce responses that are fluent, explainable, and clinically aligned.
Challenges and Design Tradeoffs
While graph RAG offers a powerful framework for grounded clinical decision support, its implementation introduces several challenges and tradeoffs—both technical and clinical. Below, we outline some of the key complexities encountered during system design and deployment.
Graph RAG vs. Vector Database Retrieval
While vector-based retrieval systems (e.g., FAISS, Weaviate, Qdrant) are widely used in LLM applications due to their speed and flexibility, they differ fundamentally from graph-based retrieval in several ways—each with distinct tradeoffs:
| Aspect | Graph RAG | Vector-Based Retrieval |
|---|---|---|
| Global Context Capture | Excels at modeling inter-entity dependencies and holistic reasoning across protocols | Often retrieves isolated snippets; lacks awareness of larger logical structure |
| Structure Awareness | Captures logical relationships, hierarchies, and causal chains | Relies on dense embedding similarity, often ignores structure |
| Query Precision | Enables exact matching of conditional paths (IF, AND, OR) |
Fuzzy matching allows generalization but may retrieve noise |
| Explainability | Transparent—retrieved paths reflect known logic | Less interpretable—matches are similarity-based and opaque |
| Flexibility | Ideal for structured data like clinical guidelines | Better suited to unstructured text like case notes or literature |
| Maintenance | Requires schema upkeep and logic versioning | Easier to update via re-embedding |
| Safety and Alignment | High: Grounded in vetted clinical logic paths; reduces hallucinations and improves traceability | Moderate: Risk of generating unsupported or out-of-context responses from semantically similar content |
Tradeoff Summary:
- Graph RAG excels when your domain has explicit structure, causality, or logic (e.g., clinical pathways, decision trees, or taxonomies). It provides grounded, explainable, and controllable retrieval—key for safety-critical tasks.
- Vector-based systems shine in large, unstructured knowledge bases, or when linguistic flexibility and broad recall matter more than logical precision.
In our IMNCI use case, where guideline logic is rule-based and highly structured, a graph-first retrieval strategy provides tighter alignment with the source of truth. However, we can often combine both approaches—using vectors for initial symptom expansion, and graphs for further logic-aware traversals, filtering, and information retrieval.
LLM Hallucination and Response Control
Even with strong grounding, LLMs are prone to hallucination—especially when prompts are loosely structured or when graph context is sparse.
- Overgeneralization: LLMs may infer treatments beyond what the graph provides if constraints aren’t explicit.
- Clinical tone and clarity: Generated text must strike a balance between accessibility for health workers and medical accuracy. Fine-tuning output formats and prompt engineering is an ongoing effort.
We mitigate these types of hallucinations by:
- Using structured prompts with explicit graph-derived facts
- Reinforcing instruction-following through examples and context scaffolding
- Post-processing responses to flag or filter unsupported claims
Static Guidelines vs. Real-World Clinical Variability
The IMNCI guidelines represent a standardized, static reference. In contrast, real-world settings introduce variability across geography, facility resources, clinician preferences, and patient populations.
- Missing localization: Some treatments or diagnostics may not be available in certain regions.
- Temporal dynamics: Follow-up recommendations depend on timing and evolving patient status—something static graphs struggle to represent.
Future work includes:
- Updating the graph with real-time variability using Bayesian techniques
- Incorporating resource constraints into the graph (e.g., “only recommend X if Y is available”)
- Extending the model to support longitudinal patient trajectories through temporal graph elements
These challenges and tradeoffs reflect the broader reality of building reliable decision support systems that bridge structured knowledge with the nuance and unpredictability of real-world care. By surfacing and addressing these tradeoffs, we aim to build safer, more adaptive tools for frontline workers.
Designing Graph Queries for Retrieval
With the IMNCI graph encoded in Neo4j, the next step is to enable dynamic querying of the data based on incoming patient observations. These queries must reflect both the structural and logical complexity of clinical reasoning—for example, determining which classifications are relevant given a combination of symptoms, or filtering treatment options based on conditional logic.
Neo4j’s Cypher query language is well-suited for this task, allowing us to traverse relationships, evaluate logical groupings, and parameterize input values.
🔍 Example 1: Retrieve Classifications for a Set of Observed Symptoms
Suppose a patient presents with two observations (symptoms): "Young infant DNA PCR
positive" and "Breastfeeding". We want to identify which set of classifications
are supported by these observations:
MATCH (c:Classification)-[:INCLUDES_OBSERVATION]->(o:Observation)
WHERE o.content IN $observedSymptoms
RETURN DISTINCT c.display_name AS ClassificationName, COUNT(*) AS MatchingObservations
ORDER BY MatchingObservations DESC
Parameter set:
This query will then rank the most likely classifications based on overlapping observations, offering a first-level diagnostic suggestion.
🔍 Example 2: Handling Conditional Logic (AND, IF, OR)
Many nodes in the IMNCI graph encode compound logic. For example, the classification
HIV STATUS UNKNOWN requires both of these to be true:
- Mother not tested
- Young infant not tested
We represent such cases using LogicGroup nodes, which model compound clinical logic
such as AND, OR, and IF. To support full-context reasoning, we often need to
retrieve nested logic blocks that lead up to a classification—for instance, when
evaluating whether all required symptoms have been met for a particular medical
classification.
The following Cypher query uses APOC procedures
to recursively collect all LogicGroup nodes connected to a set of classification
nodes:
UNWIND $classification_node_ids AS starting_node_id
CALL apoc.cypher.run(
'
MATCH (n:Classification {classification_id: $node_id})<-[:HAS_CLASSIFICATION]-(lg:LogicGroup)
OPTIONAL MATCH (lg)-[:BELONGS_TO_LOGIC_GROUP*0..]->(parentLG:LogicGroup)
RETURN $node_id AS starting_node_id,
elementId(parentLG) AS parentLG_id,
properties(parentLG) AS logic_group_properties
',
{ node_id: starting_node_id }
) YIELD value
RETURN value.parentLG_id AS id,
value.starting_node_id AS starting_node_id,
value.logic_group_properties AS logic_group_properties
This query ensures that we retrieve the complete hierarchy of logic conditions associated with each classification. Once the logic conditions are retrieved, subsequent queries can then be executed to retrieve the set of observations under each classification. This is essential when performing logic-aware filtering and downstream reasoning during patient diagnosis using the retrieved results.
🔍 Example 3: Dynamic Patient Context with Parameters
In practice, every patient’s context is different—age group, breastfeeding status, lab test availability—all factor into diagnostic decisions. To make our queries adaptive, we can expose key attributes as parameters:
MATCH (c:Classification)-[:APPLICABLE_TO]->(a:AgeGroup)
WHERE a.name = $ageGroup
WITH c
MATCH (c:Classification)-[:INCLUDES_OBSERVATION]->(o:Observation)
WHERE o.name IN $observedSymptoms
RETURN c.name AS ClassificationName
Parameter set:
{
"ageGroup": "Birth - 2 Months",
"observedSymptoms": [
"Mother not tested",
"Young infant not tested"
]
}
This flexibility allows us to use the same query templates across multiple diagnostic scenarios, while ensuring clinical relevance is preserved based on the patient's age and context.
By embedding conditional logic and domain structure directly into the graph, and querying it with parameterized Cypher templates, we create a highly adaptive retrieval layer. This layer forms the bridge between structured clinical logic and natural language generation—enabling powerful downstream applications in graph-based RAG.
Retrieval-Augmented Generation with Graph Context
Once relevant clinical information is retrieved from the IMNCI graph, the next challenge is making that information usable for LLMs in a diagnostic setting. Our approach integrates the precision of graph traversal with the natural language fluency of generative models; i.e., graph RAG. This architecture enables LLMs to reason over structured, verified content rather than relying solely on parametric memory or unstructured prompts.
Combining Full-Text/Vector and Graph-based Retrieval
While the graph structure captures rule-based medical logic with high fidelity, real-world clinical input is often unstructured, imprecise, or colloquial. To bridge that gap, we can pair graph queries with full-text/vector-based retrieval using embedding models such as:
This dual-retrieval approach allows us to:
- Semantically match user inputs to relevant clinical observations
- Retrieve and rank candidate
Observationnodes in the graph based on similarity - Use matched nodes to seed graph queries that follow logical paths (e.g., symptoms -> classifications -> treatments)
This architecture leverages the broad semantic coverage of embeddings and the structure-aware precision of graph traversal, ensuring broader language coverage without sacrificing logical integrity.
Workflow Summary: Hybrid Graph RAG in Action
Our case study below (HIV Diagnosis with Graph RAG and LLMs) illustrates this workflow in a real diagnostic scenario:
- Free-text input is embedded and compared against known
Observationnodes in the graph. - Top-matching nodes are used to anchor graph queries that traverse to
Classificationnodes according to IMNCI protocol logic. - The resulting subgraph is serialized into structured input for the LLM, which then generates a grounded, context-aware response tailored to the physician's query.
This process enables a scalable, logic-informed, and clinician-facing interface where natural language inputs can be safely mapped to actionable, evidence-based guidance.
Case Study: HIV Diagnosis with Graph RAG and LLMs
To illustrate how our graph RAG system supports real-world clinical workflows, consider the following diagnostic scenario involving HIV exposure in an infant.
A physician is in the process of assessing a newborn and has collected a small set of initial observations. In an ideal setting—with ample time, internet access, and diagnostic equipment—the physician might record a comprehensive clinical history. However, in practice, especially in low-resource environments, only limited information is often available.
In this example, the physician provides the following free-text input:
Physician: "Mother is reporting positive HIV test. Unsure about patient's PCR results."
Our graph RAG pipeline begins by interpreting this partial, unstructured input. The
goal is to identify a set of likely classifications (e.g., HIV EXPOSED,
HIV INFECTED) that are logically supported by the provided observations.
Step 1: Candidate Classification Retrieval via Full-Text Search and Graph Traversal
We first perform a full-text search against indexed Observation nodes in the Neo4j
graph, using the physician’s input as the query text. The results are used to traverse
the graph and find connected Classification nodes via their associated logic groups.
CALL db.index.fulltext.queryNodes($index_name, $query_text, $options) YIELD node AS obs
MATCH (obs)-[:BELONGS_TO_LOGIC_GROUP*]->(candidate:LogicGroup)
MATCH (candidate)-[:HAS_CLASSIFICATION]->(classification:Classification)
RETURN DISTINCT elementId(classification) AS id, labels(classification) AS labels, properties(classification) AS properties
Parameter set:
{
"index_name": "observation",
"options": {"limit": 5},
"query_text": "mother is reporting positive HIV test. unsure about patient's PCR results."
}
The query returns a focused list of relevant classifications (certain attributes are excluded for brevity):
[
{
"node_properties": {
"display_name": "HIV EXPOSED",
"classification_id": 387
}
},
{
"node_properties": {
"display_name": "HIV INFECTION UNLIKELY",
"classification_id": 389
}
},
{
"node_properties": {
"display_name": "TB DISEASE",
"classification_id": 493
}
},
{
"node_properties": {
"display_name": "HIV INFECTED",
"classification_id": 386
}
}
]
From a graph of ~90 Observation nodes and ~33 Classification nodes, this retrieval
process successfully narrows the scope to just the 4 classifications most relevant
to the physician’s scenario. Notably, classifications such as HIV STATUS UNKNOWN are
also excluded, as the available input observations logically disqualify them.
Step 2: LLM-Guided Answer Generation Using Structured Context
Next, we use the retrieved classifications to generate a context-aware, grounded response with an LLM. Because the classifications were derived from logic-aware traversal, we can provide the LLM with just the necessary and accurate context— reducing hallucination risk.
We send the retrieved classifications, the physician's query, and a guiding system prompt to a question-answering endpoint and receive the following response (certain details are excluded for brevity):
{
"answer_response": {
"explanation": "The physician is concerned about a patient whose mother has tested positive for HIV and is unsure about the patient's PCR results. The most relevant results are those that address HIV exposure in infants, the significance of PCR testing, and potential complications. Result 0, 'HIV EXPOSED', directly addresses infants born to HIV-positive mothers and the importance of PCR testing. Result 9, 'HIV INFECTED', discusses confirmed HIV infection via PCR and the need for immediate intervention. Result 3, 'HIV INFECTION UNLIKELY', is relevant as it describes scenarios where HIV infection is unlikely, providing a contrast. Result 2, 'LOW BIRTH WEIGHT AND/OR PRETERM', is also relevant because it mentions maternal HIV-positive status and pending PCR results in the context of low birth weight and preterm infants, a common comorbidity. Result 11, 'HIV STATUS UNKNOWN', is relevant as it highlights the importance of testing when the HIV status of both mother and infant is unknown. These results provide a comprehensive overview of HIV exposure, testing, and potential outcomes in infants, allowing the physician to make informed decisions.",
"answer": "The infant is classified as 'HIV EXPOSED' due to the mother's positive HIV status. A PCR test is crucial to determine the infant's HIV status. If the PCR test is positive, the infant is classified as 'HIV INFECTED' and requires immediate antiretroviral therapy (ART) and prophylactic treatment. If the PCR test is negative and the infant shows no signs of HIV infection, the classification is 'HIV INFECTION UNLIKELY'. Low birth weight and prematurity are common comorbidities in infants born to HIV-positive mothers, and their HIV status is often pending PCR testing. If both the mother and infant's HIV status are unknown, testing is urgently needed."
},
"relevant_search_results": [...],
}
Outcome
The final output offers a medically accurate, explainable response that incorporates structured clinical logic while maintaining fluency and usability for frontline providers. This case highlights how graph RAG can power real-time, logic-aware decision support grounded in trusted medical protocols—even with partial and imprecise input.
What's Next for Patient Diagnosis
You may have noticed that the diagnosis workflow illustrated above is reactive—the physician provides an open-ended query, and the system returns a structured, grounded response. However, this flow places the burden of navigation on the physician, who must know which questions to ask and how to interpret results.
Given the complexity of the IMNCI guidelines—which include dozens of diagnostic questions, clinical observations, procedure steps, classifications, and treatment plans—it can be daunting for a frontline health worker to step through the full protocol while simultaneously managing patient care.
What’s needed is a more collaborative, guided diagnostic process, where an intelligent agent (powered by an LLM) walks the physician through a structured sequence of context-aware steps. Each step would be grounded in the IMNCI graph and tailored to the patient’s age group, symptoms, and clinical presentation—ultimately converging on the most accurate diagnosis and corresponding treatment pathway.
Toward a Diagnostic Agent for Guided Clinical Reasoning
To achieve this, our next phase of development involves designing an agentic LLM workflow—a dialogue-capable assistant that can:
- Dynamically select the next best question or assessment step based on prior inputs and graph state
- Traverse the IMNCI graph interactively, following logical operators (
AND,OR,IF) to eliminate irrelevant branches - Reason about uncertainty, prompting for missing observations when multiple classifications are still plausible
- Incorporate resource constraints, adapting flows when certain procedures, tests, or medications are unavailable
- Generate interim explanations, helping physicians understand why certain questions or treatments are being recommended
This guided approach transforms patient diagnosis from a static reference lookup into an interactive reasoning experience—one that supports clinicians in real time while preserving the transparency and fidelity of structured clinical logic.
Thanks for reading! If you're working on retrieval-augmented clinical tools or building with Neo4j and LLMs, we'd love to hear from you!
⬅️ Previous: Modeling Medical Guidelines as Interactive Graphs
➡️ Next up: Managing Agentic Flows with Pydantic Graph