Modeling Medical Guidelines as Interactive Graphs
📚 View all posts in the Graph-based Healthcare Series
Graph-based Healthcare Series — 1
This is the first post in an ongoing series on graph-based healthcare tools. Stay tuned for upcoming entries on clinical modeling, decision support systems, and graph-powered AI assistants.
The Integrated Management of Neonatal and Childhood Illness (IMNCI) guidelines are a vital resource for diagnosing and treating pediatric conditions in low-resource settings. However, their traditional format—dense tables and long blocks of text—is difficult to navigate in fast-paced, high-pressure clinical environments.
To make these guidelines more usable, we built an interactive, graph-based model of the IMNCI protocol. By translating non-linear diagnostic logic into a structured, visual format, we enable faster, more intuitive decision-making and pave the way for intelligent clinical tools.
From Tables and Free-Form Text to Graphs: Why Structure Matters
Reframing medical guidelines as graph structures allows us to:
- Model clinical logic in a machine-readable format
- Visualize relationships between symptoms, classifications, and actions
- Enable step-by-step navigation for decision-making
- Support automation in medical AI or decision-support tools
This structured representation unlocks new possibilities for training, inference, and deployment of intelligent clinical systems.
Step 1. Extracting Structured Data from Clinical Tables and Free-Form Text
To build the foundation of our graph model, we first extract structured data from the IMNCI guidelines. Each guideline contains components such as:
- Diagnostic questions
- Clinical signs (observed symptoms)
- Medical classifications
- Treatment plans
- Follow-up guidance
- Clinical notes and definitions
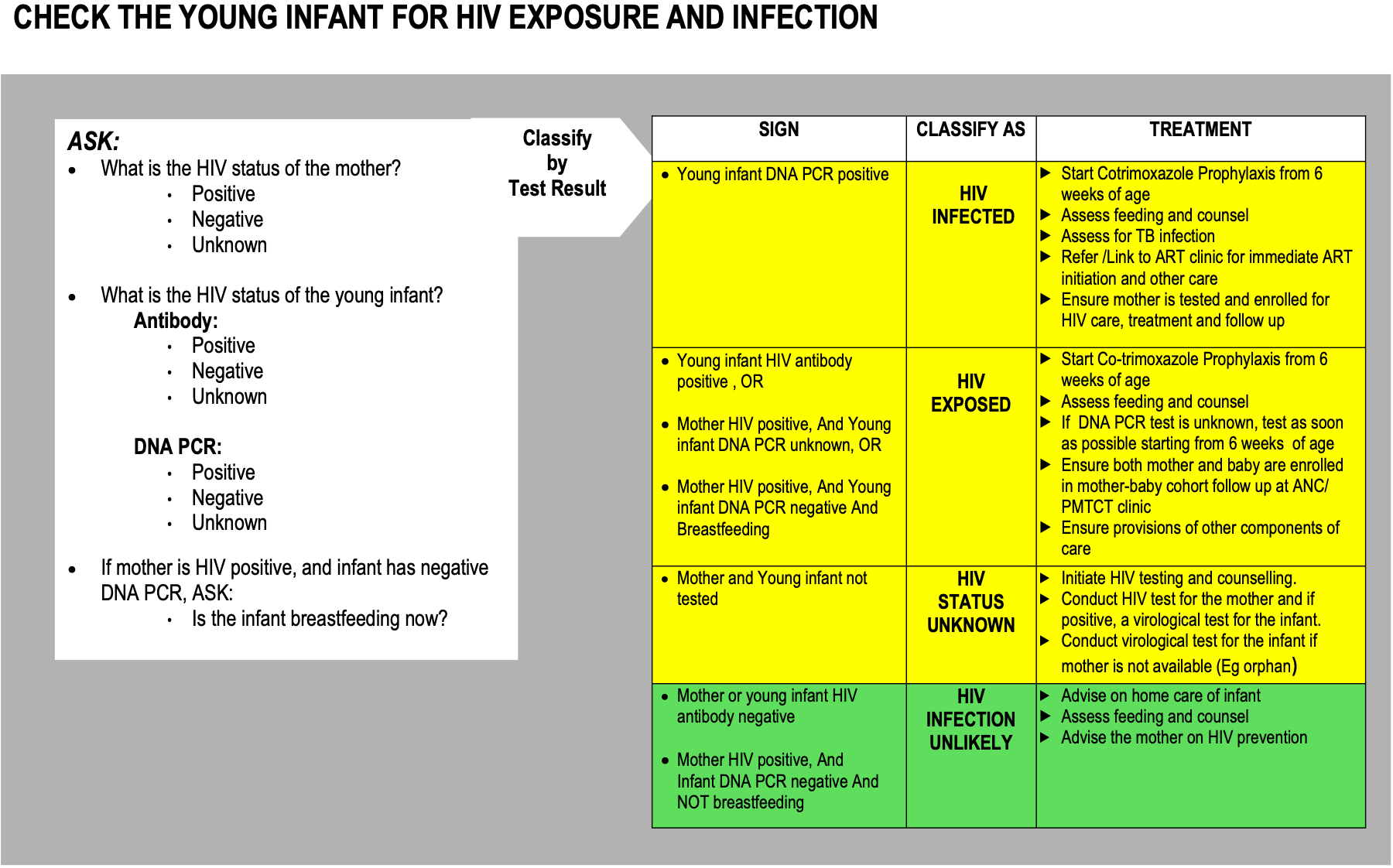
Below is an excerpt from the IMNCI guideline for diagnosing and treating HIV Exposure and Infection.
Information Extraction Using Vision-Language Models
We used vision-language models (VLMs) that process both text and visual input to convert screenshots of these guidelines into the following structured JSON format:
{
"Age Group": {
"Medical Condition": {
"AFTER": {...},
"ASSESSMENT": {...},
"BEFORE": {...},
"CLASSIFICATION": {...},
"CONTEXT": "...",
"CRITICAL_INFORMATION": {...},
"FOOTNOTE": {...},
"OBSERVATION": {...},
"PROCEDURE": {...},
"TREATMENT": {...}
}
}
}
Mapping JSON Keys to Clinical Logic
Each key in the JSON maps to a specific part of the guideline's logic or action as follows:
Age Group: The age range applicable to the guideline. For IMNCI data, this corresponds to eitherBirth - 2 Monthsor2 Months - 5 Years.Medical Condition: The specific medical condition being addressed, such asHIV Exposure and Infection,Pneumonia,Birth Asphyxia, etc.- For each medical condition, the following sub-keys are present:
AFTER: Related medical conditions that should be evaluated following the current classification.ASSESSMENT: Details on how to assess the condition.BEFORE: Related medical conditions that should be evaluated prior to the current classification.CLASSIFICATION: The medical classification(s) related to the condition.CONTEXT: Additional context or notes related to the condition.CRITICAL_INFORMATION: Important information that must be considered.OBSERVATION: Observations/signs/symptoms related to each classification associated with the condition.PROCEDURE: Procedures to follow when diagnosing the condition.TREATMENT: Treatment plans related to each classification associated with the condition.
Click to view full JSON example for 'HIV Exposure and Infection'
{
"HIV Exposure and Infection": {
"AFTER": {
"YELLOW 1": {
"AND": [
{
"IF": {
"Breastfeeding": {
"AND": [
"Feeding Problem or Underweight - Breastfeeding"
]
}
}
},
{
"IF": {
"Not breastfeeding": {
"AND": [
"Feeding Problem or Underweight - Not Breastfeeding"
]
}
}
},
"Tuberculosis"
]
},
"YELLOW 2": {
"AND": [
{
"IF": {
"Breastfeeding": {
"AND": [
"Feeding Problem or Underweight - Breastfeeding"
]
}
}
},
{
"IF": {
"Not breastfeeding": {
"AND": [
"Feeding Problem or Underweight - Not Breastfeeding"
]
}
}
}
]
},
"GREEN 1": {
"AND": [
{
"IF": {
"Breastfeeding": {
"AND": [
"Feeding Problem or Underweight - Breastfeeding"
]
}
}
},
{
"IF": {
"Not breastfeeding": {
"AND": [
"Feeding Problem or Underweight - Not Breastfeeding"
]
}
}
}
]
}
},
"ASSESSMENT": {
"AND": [
"What is the HIV status of the mother?\n\t\u2022 Positive\n\t\u2022 Negative\n\t\u2022 Unknown",
"What is the HIV status of the young infant?\n\tAntibody:\n\t\t\u2022 Positive\n\t\t\u2022 Negative\n\t\t\u2022 Unknown\n\tDNA PCR:\n\t\t\u2022 Positive\n\t\t\u2022 Negative\n\t\t\u2022 Unknown",
{
"IF": {
"Mother is HIV positive and infant has negative DNA PCR": {
"AND": [
"Ask if the infant is breastfeeding now"
]
}
}
}
]
},
"BEFORE": null,
"CLASSIFICATION": {
"YELLOW 1": "HIV INFECTED",
"YELLOW 2": "HIV EXPOSED",
"YELLOW 3": "HIV STATUS UNKNOWN",
"GREEN 1": "HIV INFECTION UNLIKELY"
},
"CONTEXT": "Checking the young infant for HIV exposure and infection.",
"CRITICAL_INFORMATION": null,
"FOOTNOTE": null,
"OBSERVATION": {
"YELLOW 1": {
"AND": [
"Young infant DNA PCR positive"
]
},
"YELLOW 2": {
"AND": [
{
"OR": [
"Young infant HIV antibody positive",
{
"AND": [
"Mother HIV positive",
"Young infant DNA PCR unknown"
]
},
{
"AND": [
"Mother HIV positive",
"Young infant DNA PCR negative",
"Breastfeeding"
]
}
]
}
]
},
"YELLOW 3": {
"AND": [
"Mother not tested",
"Young infant not tested"
]
},
"GREEN 1": {
"AND": [
{
"OR": [
"Mother or young infant HIV antibody negative",
{
"AND": [
"Mother HIV positive",
"Infant DNA PCR negative",
"NOT breastfeeding"
]
}
]
}
]
}
},
"PROCEDURE": null,
"TREATMENT": {
"YELLOW 1": {
"Urgent pre-referral": null,
"Other": {
"AND": [
"Start Co-trimoxazole Prophylaxis from 6 weeks of age",
"Assess feeding and counsel",
"Assess for TB infection",
"Refer/Link to ART clinic for immediate ART initiation and other care",
"Ensure mother is tested and enrolled for HIV care, treatment and follow up"
]
}
},
"YELLOW 2": {
"Urgent pre-referral": null,
"Other": {
"AND": [
"Start Co-trimoxazole Prophylaxis from 6 weeks of age",
"Assess feeding and counsel",
{
"IF": {
"DNA PCR test is unknown": {
"AND": [
"Test as soon as possible starting from 6 weeks of age"
]
}
}
},
"Ensure both mother and baby are enrolled in mother-baby cohort follow up at ANC/PMTCT clinic",
"Ensure provisions of other components of care"
]
}
},
"YELLOW 3": {
"Urgent pre-referral": null,
"Other": {
"AND": [
"Initiate HIV testing and counselling.",
{
"IF": {
"Mother is available": {
"AND": [
"Conduct HIV test for the mother",
{
"IF": {
"Mother's HIV test is positive": {
"AND": [
"Conduct a virological test for the infant."
]
}
}
}
]
}
}
},
{
"IF": {
"Mother is not available (e.g., infant is an orphan)": {
"AND": [
"Conduct virological test for the infant"
]
}
}
}
]
}
},
"GREEN 1": {
"Urgent pre-referral": null,
"Other": {
"AND": [
"Advise on home care of infant",
"Assess feeding and counsel",
"Advise the mother on HIV prevention"
]
}
}
}
}
}
Step 2. Using Logic Structures to Decompose Complex Information
Even once structured, IMNCI guidelines contain dense, nonlinear logic. To better
navigate this complexity, we decompose the extracted information into smaller
components using logical operators such as AND, OR, IF, etc.
For instance, to classify a patient under HIV STATUS UNKNOWN, both conditions below
must be true:
The classification only applies when both observations are present.
Similarly, treatment logic often includes qualifiers:
{
"IF": {
"DNA PCR test is unknown": {
"AND": [
"Test as soon as possible starting from 6 weeks of age"
]
}
}
}
This step is conditionally applied if the DNA PCR test is unknown, using the
IFoperator to model that dependency.
Breaking down these kinds of logic allows us to:
- Create a graph structure that faithfully models dependencies and flows.
- Simplify the guideline for machine learning and decision-support tools (e.g., large language models).
Step 3. Building the Graph Model in Neo4j
To bring this structured logic to life, we built a graph model using Neo4j—a popular graph database platform.
Designing a graph model begins with defining a set of organizing principles:
- Nodes: Entities being modeled.
- Node Properties: Metadata describing each node.
- Relationships: How nodes are connected.
- Relationship Properties: Attributes of each connection.
Careful consideration must be given to the design of the graph model, as this step dictates not only how the data will be stored but also how it can be queried and used in downstream applications. Explicit definitions of relationships will enable certain queries to be more efficient, while the inclusion of certain properties will allow for more detailed analysis and insights.
Example node types include:
- Classification:
HIV INFECTED,HIV EXPOSED - Condition:
Very Severe Disease,Local Bacterial Infection - Observation:
Young infant DNA PCR positive - Treatment Step:
Start Co-trimoxazole Prophylaxis
Example relationships include:
(Condition) -[:HAS_CLASSIFICATION]-> (Classification)(Observation) -[:MIGHT_INCLUDE_OBSERVATION]-> (Condition)(Logic Group) -[:INCLUDES_TREATMENT_STEP]-> (Treatment Step)
Sample Classification properties include:
classification_id: Unique ID of the classificationdescription: Full description of the classificationdisplay_name: Human-readable label (corrected spelling)
To ingest the data, we used Python scripts to recursively transform the JSON into CSV files, which were then imported into Neo4j using Cypher queries.
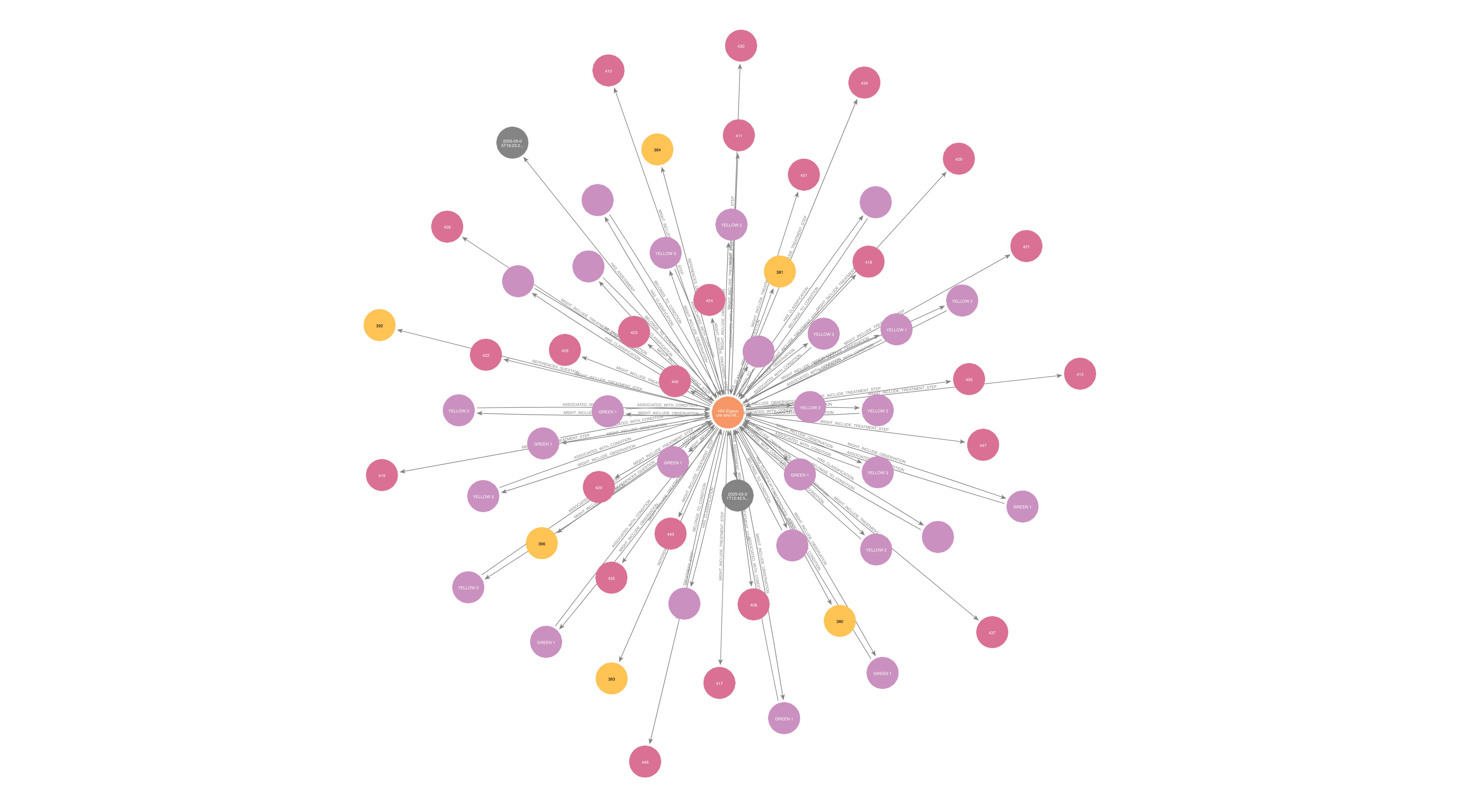
Bringing It All Together: A Visual, Navigable Knowledge Graph
This process produced over 600 nodes and 1,600 relationships per age group across the IMNCI guidelines. The resulting graph allows for both human exploration and machine processing of clinical logic.
A closer look at HIV Exposure and Infection reveals a richly interconnected view of
symptoms, classifications, and actions:
Real-World Impact and Future Directions
By transforming IMNCI guidelines into a navigable knowledge graph, we lay the groundwork for smarter clinical tools. These models can power:
- Point-of-care decision support for frontline workers
- Mobile applications that simplify diagnosis steps
- LLM-based assistants that reason over structured logic
- Clinical training tools that visualize patient flows
This graph-first approach to clinical modeling opens the door to AI systems that are not just intelligent—but also clinically aligned, transparent, and ready to scale. We're excited to continue exploring this space, and welcome feedback and collaboration from the broader clinical AI and informatics community.
Thanks for reading! If you're working on clinical AI, health informatics, or decision-making support, we'd love to hear from you!
➡️ Next up: Enhancing Patient Diagnosis with Graph-based Retrieval-Augmented Generation