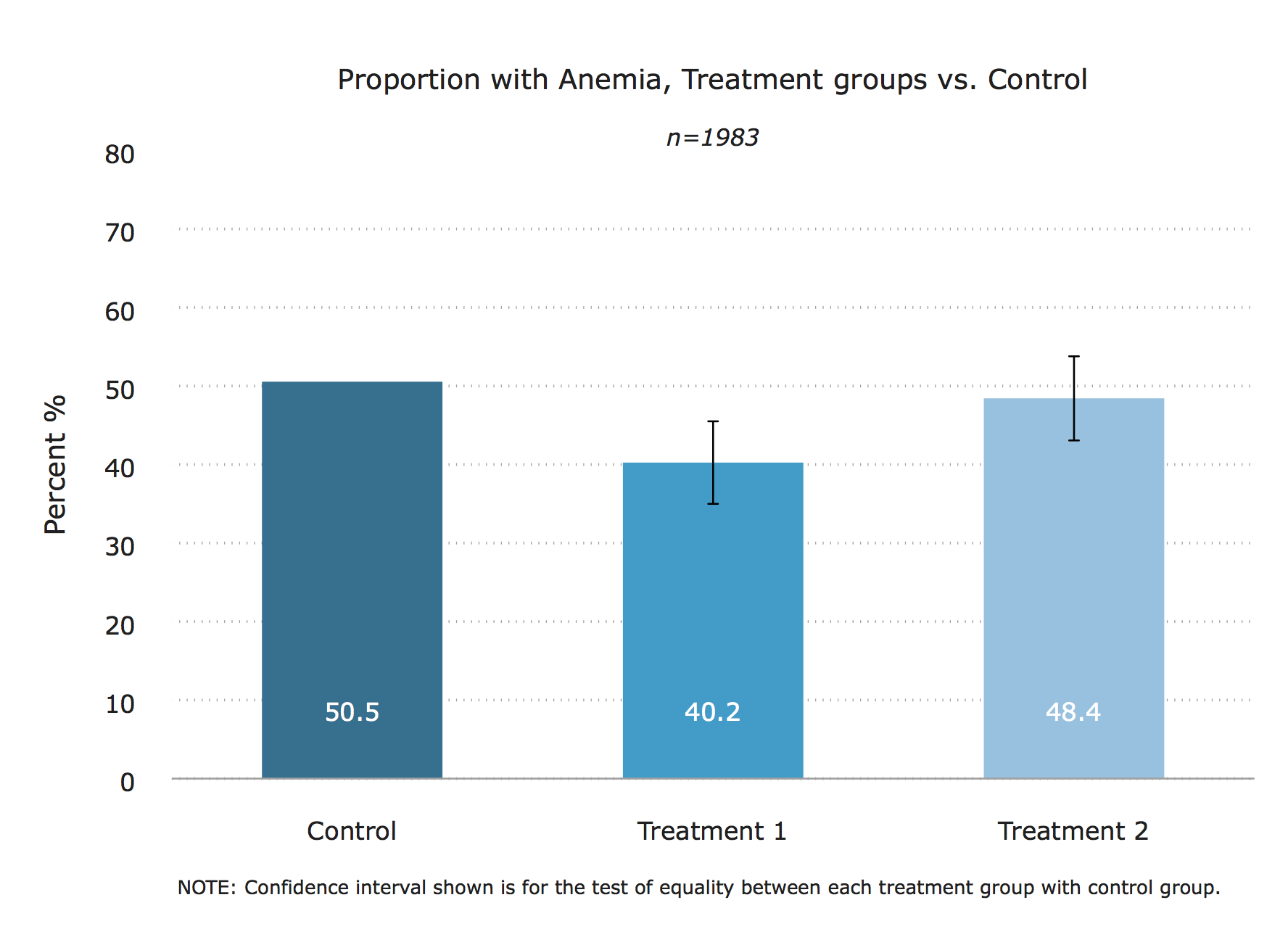
The chart
Stata
R
Data
Sample dataset in .dta format can be downloaded here. The R code uses the same dataset in .csv format, which can be found here.
The code
Stata
*_______________________________________________________________________________
* Project : IDinsight data visualization guide - Stata graph templates
*
* Graph Type : Treatment vs. control bar graphs w/ confidence interval
* for the treatment effect (TYPE 2)
* Figure displays control and treatment group means +
* confidence interval for the test of equality between each
* treatment group & control.
* By : Crystal Huang
* Last edited : May 23, 2019
*_______________________________________________________________________________
* Toy dataset
global data "/Users/crystalhuang/Dropbox (IDinsight)/Data Visualization Guide/3. Toy Data"
global output "/Users/crystalhuang/Dropbox (IDinsight)/Data Visualization Guide/2. Graph templates/graphs"
use "$data/toydata.dta", clear
* Set IDinsight scheme and font
set scheme idinsight
graph set window fontface "Verdana"
* 1. Run regression to get treatment effects for T1 and T2
reg anemic treat1 treat2
* 2. Store output in matrix- sample size for the graph later
mat A = r(table)
local samp = e(N)
* 3. Store regression results we need in matrices (to be later made into a dataset)
matlist A
* a) Estimated Treatment 1 mean (T1 effect + _cons) and CI of treatment effect
local mean_t1= A[1,1] + A[1,3]
local se_t1 = A[2,1]
mat T1= [`mean_t1', `se_t1']
mat rownames T1 = "treatment 1"
* b) Estimated Treatment 2 mean (T2 effect + _cons) and CI of treatment effect
local mean_t2 = A[1,2] + A[1,3]
local se_t2 = A[2,2]
mat T2 = [`mean_t2',`se_t2']
mat rownames T2 = "treatment 2"
* c) Adjusted Control mean (coefficient on _cons)
local c= A[1,3]
mat C= [`c',.]
matlist C
mat rownames C= "control"
* d) Vertically combine the matrices of results
mat result= T1\T2\C
mat colnames result = "mean" "se"
*4. Save matrix into a new dataset
clear
qui svmat result, names(col)
g treat= 1 in 1
replace treat= 2 in 2
replace treat= 0 in 3
* create CIs for treatment groups
g lo = mean + 1.96*se
g hi = mean - 1.96*se
*5. Format bar labels into percentages with 1 decimal place
foreach var in mean hi lo {
replace `var'= `var'*100
}
format mean %4.1f
*6. Create x-axis variable "x" that contains the order of bars that will appear in your plot
sort treat
g x= _n
* Create y-axis variable that contains y-placement for bar labels
g y= 2
#delimit;
twoway (bar mean x if treat==0, barwidth(0.5))
(bar mean x if treat==1, barwidth(0.5))
(bar mean x if treat==2, barwidth(0.5))
(rcap hi lo x, lwidth(thin) lcolor(black))
(scatter y x, msym(none) mlab(mean) mlabpos(12) mlabgap(4) mlabsize(small) mlabcolor(white)),
ylab(0(10)80, format(%4.0f))
xlab(0.5 " " 1 "Control" 2 "Treatment 1" 3 "Treatment 2" 3.5 " ", nogrid)
xtitle("")
ytitle("Percent %")
yscale(lcolor(white))
title("Proportion with Anemia, Treatment groups vs. Control")
subtitle("{it}n=`samp'{sf}")
caption("NOTE: Confidence interval shown is for the test of equality between each treatment group with control group.")
legend(off);
graph export "$output/bar_TvsC_type2.tif", replace ;
#delimit cr
R
# Treat/control V2
############################### Initial Setup ###################################
# Install required packages if they are not already in your system
packages <- c('readr',
'tidyverse')
lapply(packages, function(i) {if(!i %in% installed.packages() == T)
{install.packages(i, dependencies = TRUE, repos='http://cran.rstudio.com/')}})
# Loading required packages
library("readr")
library("tidyverse")
# Setting working directory
setwd("~/Dropbox (IDinsight)/Data visualization library")
################################ Loading dataset ###############################
mydata <- read_csv("Data/toydata.csv", show_col_types = FALSE)
############################### Data processing ################################
# Setting the right data type
# Converting the treatment variables to a factor variables
# This is because treatment will appear on the x-axis and it has only 2
# values (discrete)
mydata$treat1 <- as.factor(mydata$treat1)
mydata$treat2 <- as.factor(mydata$treat2)
# Getting the treatment effects
model <- lm(anemic ~ treat1 + treat2, data = mydata)
model_summary <- summary(model)
model_confint <- confint(model)
# Storing the sample size
samp <- nobs(model)
# Estimated Treatment 1 mean (T1 effect + intercept)
mean_t1 <- model_summary$coefficients[1, 1] + model_summary$coefficients[2, 1]
se_t1 <- model_summary$coefficients[2, 2]
# Estimate Treatment 2 mean (T2 effect + intercept)
mean_t2 <- model_summary$coefficients[1, 1] + model_summary$coefficients[3, 1]
se_t2 <- model_summary$coefficients[3, 2]
# Adjusted control mean (coefficient on intercept)
mean_control <- model_summary$coefficients[1, 1]
# Combining to a single data frame
means <- rbind(mean_t1, mean_t2, mean_control)
ses <- rbind(se_t1, se_t2, NA)
status <- c(1, 2, 0)
graph_data <- as.data.frame(cbind(status, means, ses))
rownames(graph_data) <- NULL
colnames(graph_data) <- c("treat", "mean", "se")
graph_data$treat <- as.factor(graph_data$treat)
################################ Creating the graph ############################
# Define axis and fill variable
x_values <- graph_data$treat
y_values <- graph_data$mean
plot <- graph_data %>%
# Setting aesthetic which will be inherited by other geometric objects
ggplot(aes(x = x_values, y = y_values)) +
# "identity" selected for stat since we want the heights of the bars to
# represent values in the data. The default, "bin" makes the height of each
# bar equal to the number of cases in each group.
# Fill is set according to the treatment status
# Width determines the width of the bars
geom_bar(aes(fill = treat), stat = "identity", width = 0.7) +
# Creates the error bars on top of the bars
geom_errorbar(aes(ymin = (y_values - 1.96*se), ymax = (y_values + 1.96*se)),
width = 0.06,
size = 0.5) +
# Creates the mean text on top of the error bars. It is position at y = 0.09
# units. This puts it inside the bar
geom_text(aes(y = 0.09, label = round(y_values*100, 1)), color = "white",
size = 4.5) +
# Creates the text with the n
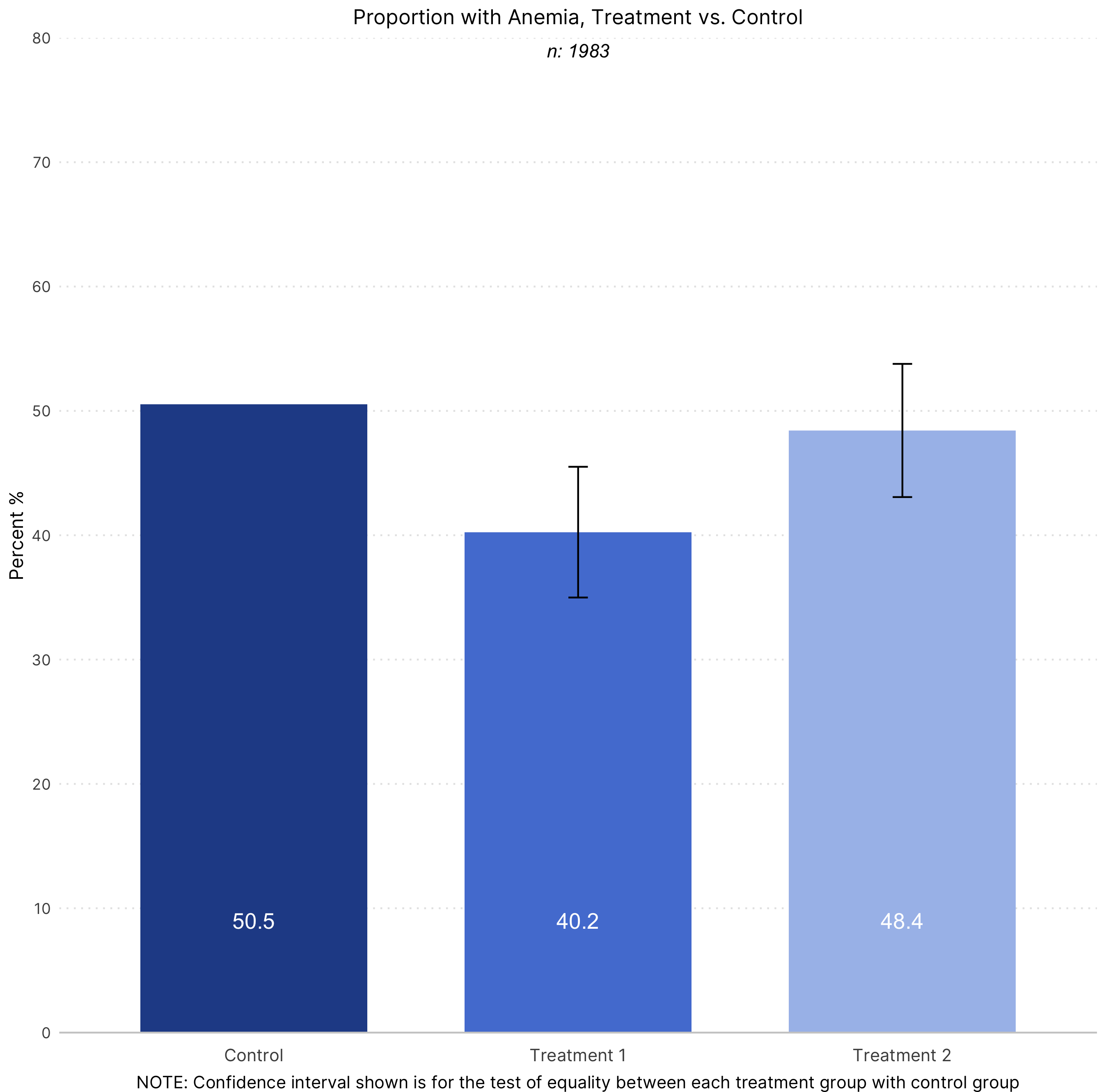
annotate(geom = "text", y = 0.79, x = 2,
label = paste0("n: ", samp),
fontface = "italic") +
# Choosing the bar colors from IDinsight brand colors
scale_fill_manual(values = c("#264D96", "#5480D6", "#A8BFEB")) +
# Expand with (0, 0) removes the space between the axis and the plot.
# Limits determine the extent of the y-axis
# Breaks determine the axis ticks
# Labels determine what is displayed at each tick
scale_y_continuous(expand = c(0, 0),
limits = c(0, 0.8),
breaks = c(seq(from = 0, to = 0.8, by = 0.1)),
labels = c("0", "10", "20", "30", "40", "50",
"60", "70", "80")) +
# Setting the x-axis labels
scale_x_discrete(breaks = c(0, 1, 2),
labels = c("Control", "Treatment 1", "Treatment 2")) +
# Graph, axis title and caption labels
labs(title = "Proportion with Anemia, Treatment vs. Control",
y = "Percent %",
caption = "NOTE: Confidence interval shown is for the test of equality between each treatment group with control group")
############################# Formatting the graph #############################
plot +
theme_classic() +
# The following visual elements have been modified:
# Set the overall font as Inter
# Removed the x-axis title
# Set the x-axis line as a light shade of gray
# Removed the y-axis line
# Create y-axis grid lines which are light gray and dotted
# Removed axis ticks
# Removed the legend
# Vertically aligned and resized the x-axis text
# Horizontally and vertically aligned and resized the caption
# Horizontally aligned and resized the graph title
theme(text = element_text(family = "Inter"),
axis.title.x = element_blank(),
axis.line.x = element_line(color = "grey80"),
axis.line.y = element_blank(),
panel.grid.major.y = element_line(colour = "grey90", linetype = "dotted"),
axis.ticks = element_blank(),
legend.position = "none",
axis.text.x = element_text(vjust = -1, size = 10),
plot.caption = element_text(hjust = 0.5, vjust = -3, size = 10),
plot.title = element_text(hjust = 0.5, size = 12))
############################# Saving and exporting #############################
#indicating the export folder and the image file name
export_folder <- "R/Bar graphs/Exports/"
img_name <- "bar_treat_control_v2_R_reviewed.png"
ggsave(paste(export_folder,img_name,sep = ""))
Other details
Stata
Credit: Crystal Huang
You must have IDinsight styles installed to get replicate the chart above.
R
Code written by Arkadeep Bandyopadhyay and reviewed by Sandra Alemayehu.
Colors for the graph have been chosen from IDinsight’s brand guide.