The chart
Stata
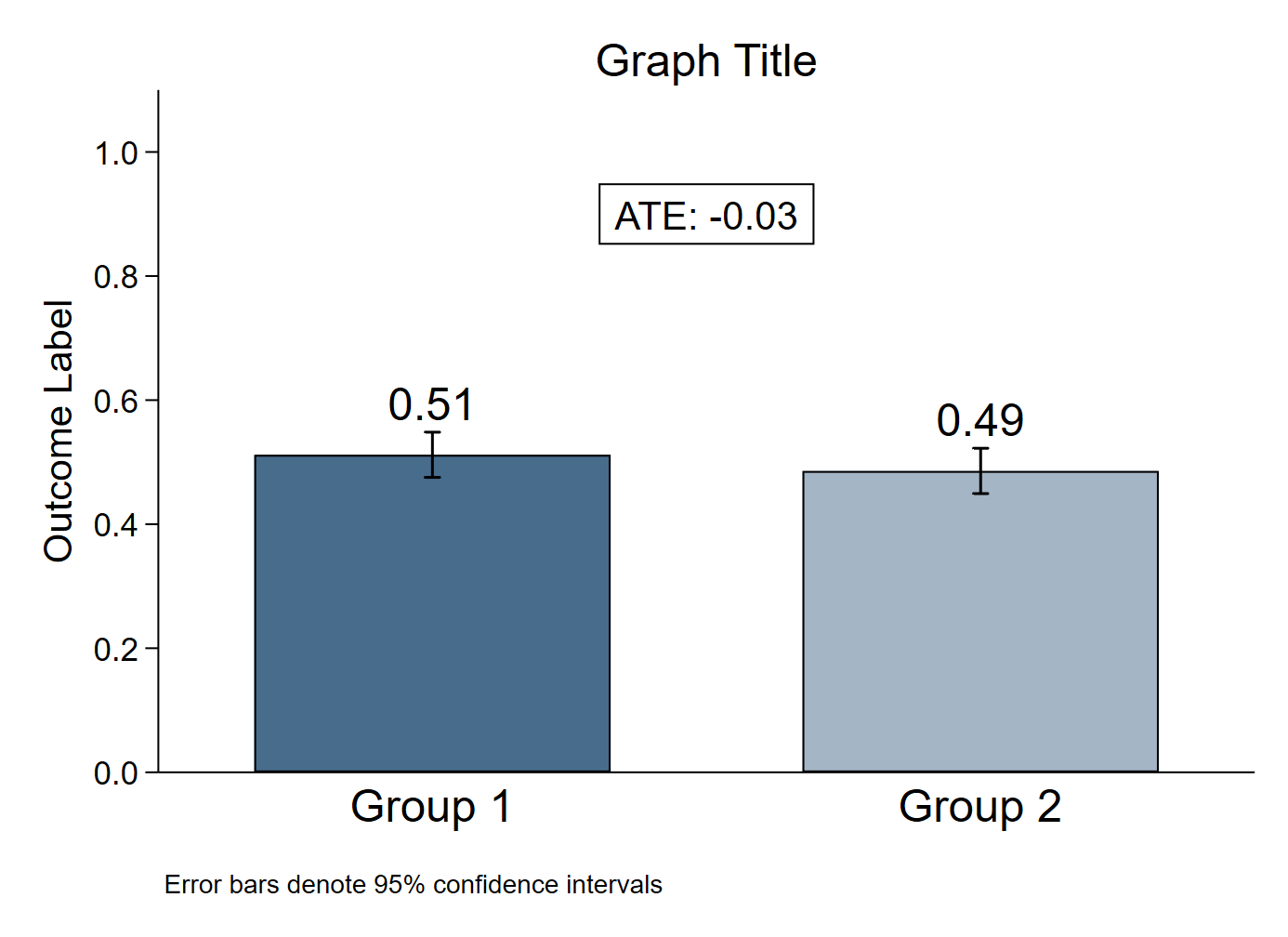
R
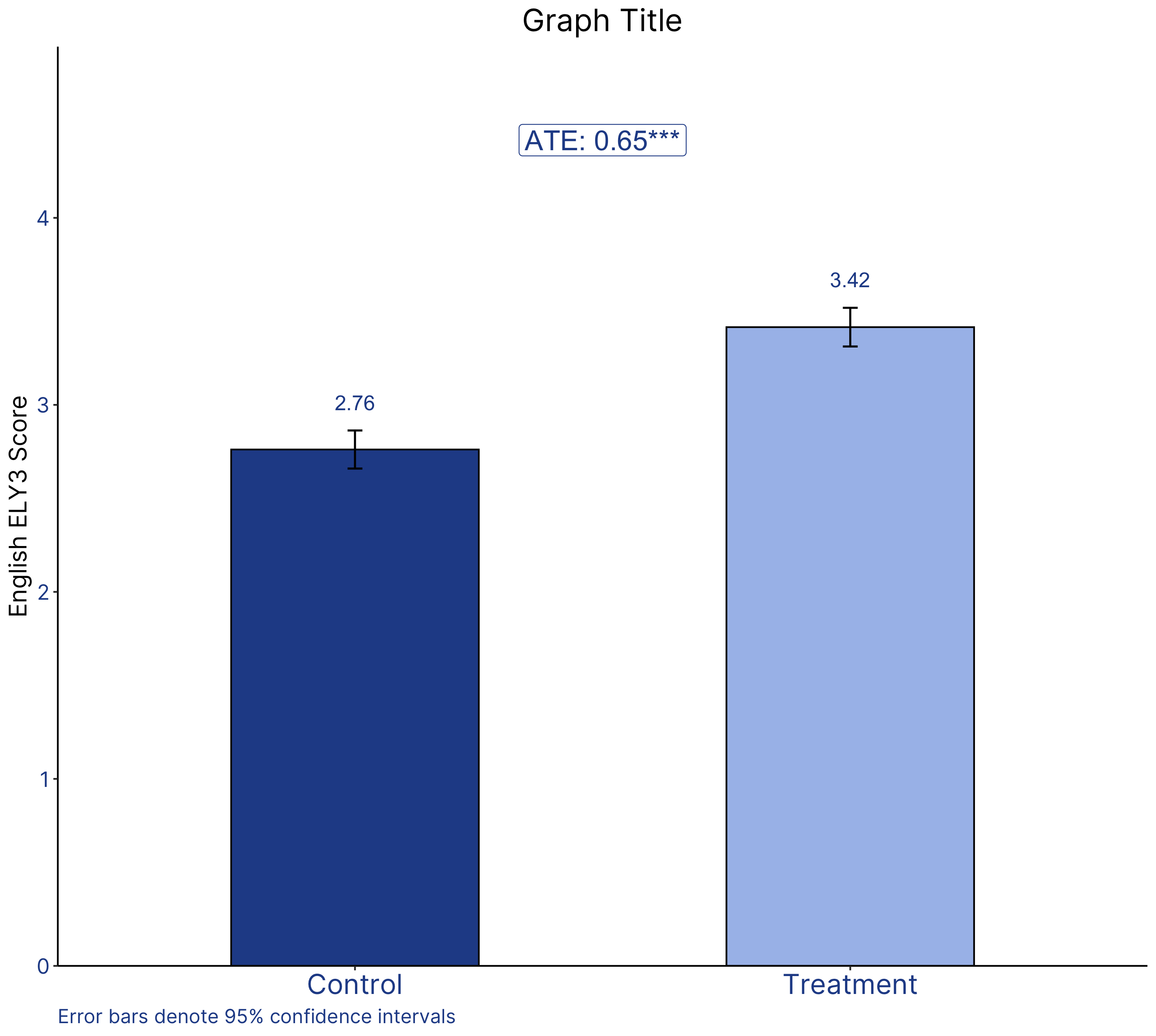
The R graph looks different since it has been created using a different dataset.
Data
Dataset used to create the R version of the graph can be found here.
The code
Stata
* Create fake dataset (delete this section and import your own data)
clear
set obs 500
gen outcome_var = runiform()
gen group_var = (_n > 0.5*_N)
* Enter your relevant variable names here
local outcome outcome_var
local group group_var
* Change the graph font here if you want
// graph set window fontface "Times New Roman"
* Create variables for graph
gen xaxis = _n in 1/2
gen means = .
gen ci_high = .
gen ci_low = .
* Calculate means, CIs, ATE
* Group 1 values
mean `outcome' if `group' == 0
matrix A = r(table)
replace means = A[1,1] in 1
replace ci_low = A[5,1] in 1
replace ci_high = A[6,1] in 1
* Group 2 values
mean `outcome' if `group' == 1
matrix A = r(table)
replace means = A[1,1] in 2
replace ci_low = A[5,1] in 2
replace ci_high = A[6,1] in 2
* ATE
reg `outcome' `group', robust
matrix A = r(table)
local ate = A[1,1]
local ate: disp %3.2f `ate'
local pval = A[4,1]
if `pval' < 0.1{
local pval_stars *
}
if `pval' < 0.05{
local pval_stars **
}
if `pval' < 0.01{
local pval_stars ***
}
* Format labels for bar heights
format %3.2f means
* Graph
twoway (bar means xaxis if xaxis == 1, fcolor(navy) lcolor(black) barwidth(0.65)) ///
(bar means xaxis if xaxis == 2, fcolor(navy*0.5) lcolor(black) barwidth(0.65)) ///
(rcap ci_high ci_low xaxis, lcolor(black)) ///
(scatter ci_high xaxis, msymbol(none) mlabel(means) mlabsize(large) mlabcolor(black) mlabposition(12)) ///
, ///
text(0.9 1.5 "ATE: `ate'`pval_stars'", box fcolor(none) lcolor(black) size(medlarge) margin(small)) ///
xtitle(" ") ///
xlabel(1 "Group 1" 2 "Group 2", labsize(large) notick) ///
xscale(range(0.5 2.5)) ///
ylabel(0(0.2)1, format(%3.1f) angle(horizontal)) ///
yscale(range(0 1.1)) ///
ytitle("Outcome Label", size(medlarge)) ///
title("Graph Title") ///
note("Error bars denote 95% confidence intervals") ///
legend(off) ///
scheme(s1color) ///
plotregion(margin(zero) style(none))
graph export "Bar graph with CIs_2 bars.png", replace
R
# Bar graph with CIs - 2 groups
################################ Initial Setup #################################
# Install required packages if they are not already in your system
packages <- c('tidyverse')
lapply(packages, function(i) {if(!i %in% installed.packages() == T)
{install.packages(i, dependencies = TRUE, repos='http://cran.rstudio.com/')}})
# Loading required packages
library("tidyverse")
# Setting working directory
setwd("~/Dropbox (IDinsight)/Data visualization library")
############################## Loading dataset #################################
mydata <- read_csv("Data/EG_DIB.csv")
############################## Data processing #################################
##>>> Setting correct data type
# Converting the treatment variable to a factor(categorical) variable
# This is because treatment will appear on the x-axis and it has only 2
# values (discrete)
mydata$treatment <- as.factor(mydata$treatment)
# Creating the means, CIs and ATE
# Assuming 95% CI
alpha = 0.05
# Creating means and CI and storing in my_sum tibble
my_sum <- mydata %>%
group_by(treatment) %>%
summarise(n = n(),
mean = mean(english_ely3_villavg, na.rm = T),
sd = sd(english_ely3_villavg, na.rm = T)
) %>%
mutate(se = sd/sqrt(n)) %>%
mutate(ic = se * qt((1-alpha)/2 + 0.5, n - 1))
# Obtaining the value of ATE and corresponding p value
model <- lm(english_ely3_villavg ~ treatment, data = mydata)
ate <- summary(model)$coefficients[2,1]
p_value <- summary(model)$coefficients[2,4]
# Creating a variable to store the significance stars
if(summary(model)$coefficients[2,4] < 0.01) {
p_stars = "***"
} else if (summary(model)$coefficients[2,4] < 0.05) {
p_stars = "**"
} else if (summary(model)$coefficients[2,4] < 0.1) {
p_stars = "*"
} else {
p_stars = ""
}
############################## Creating the graph ##############################
# Define axis and fill variable
x_values <- my_sum$treatment
y_values <- my_sum$mean
fill_by <- my_sum$treatment
plot1 <- ggplot(my_sum) +
# Geometric object for the bars
# fill inside the aes function determines the color inside the bar. Here the
# color changes based on the value of fill_by (treatment)
# color outside of the aes function determines the outline color. Here we have
# set it to black
# "identity" selected for stat since we want the heights of the bars to
# represent values in the data. The default, "bin" makes the height of each
# bar equal to the number of cases in each group.
# width determines the width of the bars
geom_bar(aes(x = x_values, y = y_values, fill = fill_by),
color = "black",
stat = "identity",
width = 0.5) +
# Manually coloring the two bars according to IDinsight colors
scale_fill_manual(values = c("#264D96", "#A8BFEB")) +
# Manually labeling the x-axis ticks for the two bars
# scale_x_discrete instead of continuous because the treatment variable
# is a factor
scale_x_discrete(breaks = c(0, 1),
labels = c("Control", "Treatment")) +
# The y-axis is continuous since the the English scores can have values such
# as 1.5, 3.8, etc.
# The expand function provides padding around the data so that it is presented
# some distance away from the axes. In this case, we are removing the padding.
# The limits function is providing the range of data to be displayed on the
# y-axis. Here, we are starting from 0 and ending it 1.5 units above the
# maximum value of the mean English end line 3 score. We are doing this to
# ensure that the ATE box has enough vertical space to be visible.
scale_y_continuous(expand = c(0, 0),
limits = c(0, max(y_values) + 1.5)) +
# Geometric object for adding the error bars
geom_errorbar(aes(x = treatment, ymin = y_values - ic, ymax = y_values + ic),
width = 0.03,
size = 0.6) +
# Geometric object for adding the mean of the bars on top of the upper bound
# of the CI
# The y value of (y_values + ic + 0.15) indicates that the text should appear
# 0.15 units above (y_values + ic)
geom_text(aes(x = x_values, y = y_values + ic + 0.15,
label = round(y_values, 2)), size = 4.5, color = "#264D96") +
# Annotate lets us to place the ATE box.
# The y-coordinate is placed 1 unit above the maximum value of the means
# The x-coordinate is placed in the center of the two bars 1 and 2 and
# hence takes the value 1.5
# paste0 is a function which concatenates strings and objects
annotate(geom = "label", x = 1.5, y = (max(y_values) + 1),
label = paste0("ATE: ", round(ate, digits = 2), p_stars),
size = 6, color = "#264D96") +
# Creating titles for the graph
labs(title = "Graph Title",
y = "English ELY3 Score",
caption = "Error bars denote 95% confidence intervals")
############################ Formatting the graph ##############################
plot1 +
# Classic theme removes grid lines and background
theme_classic() +
# Custom theme elements will come AFTER a pre-built theme has been applied
# The following changes have been made:
# Applied the Inter font to all text elements on the graph
# Removed x-axis title
# Increased the size the of y-axis title
# Changed the color and font size of the x-axis labels
# Changed the color and font size of the y-axis labels
# Removed the legend
# Center aligned the graph title and increased its size
# Left aligned the caption and changed caption color
theme(text = element_text(family = "Inter"),
axis.title.x = element_blank(),
axis.title.y = element_text(size = 15),
axis.text.x = element_text(size = 17, color = "#264D96"),
axis.text.y = element_text(size = 13, color = "#264D96"),
legend.position = "none",
plot.title = element_text(hjust = 0.5, size = 19),
plot.caption = element_text(hjust = 0, colour = "#264D96",
size = 12))
########################## Saving and exporting ################################
#indicating the export folder and the image file name
export_folder <- "R/Bar graphs/Exports/"
img_name <- "bar_graph_ci_R_reviewed.png"
ggsave(paste(export_folder,img_name,sep = ""))
Other details
R
Code written by Arkadeep Bandyopadhyay and reviewed by Sandra Alemayehu.
Colors for the graph have been selected from IDinsight’s brand guide.