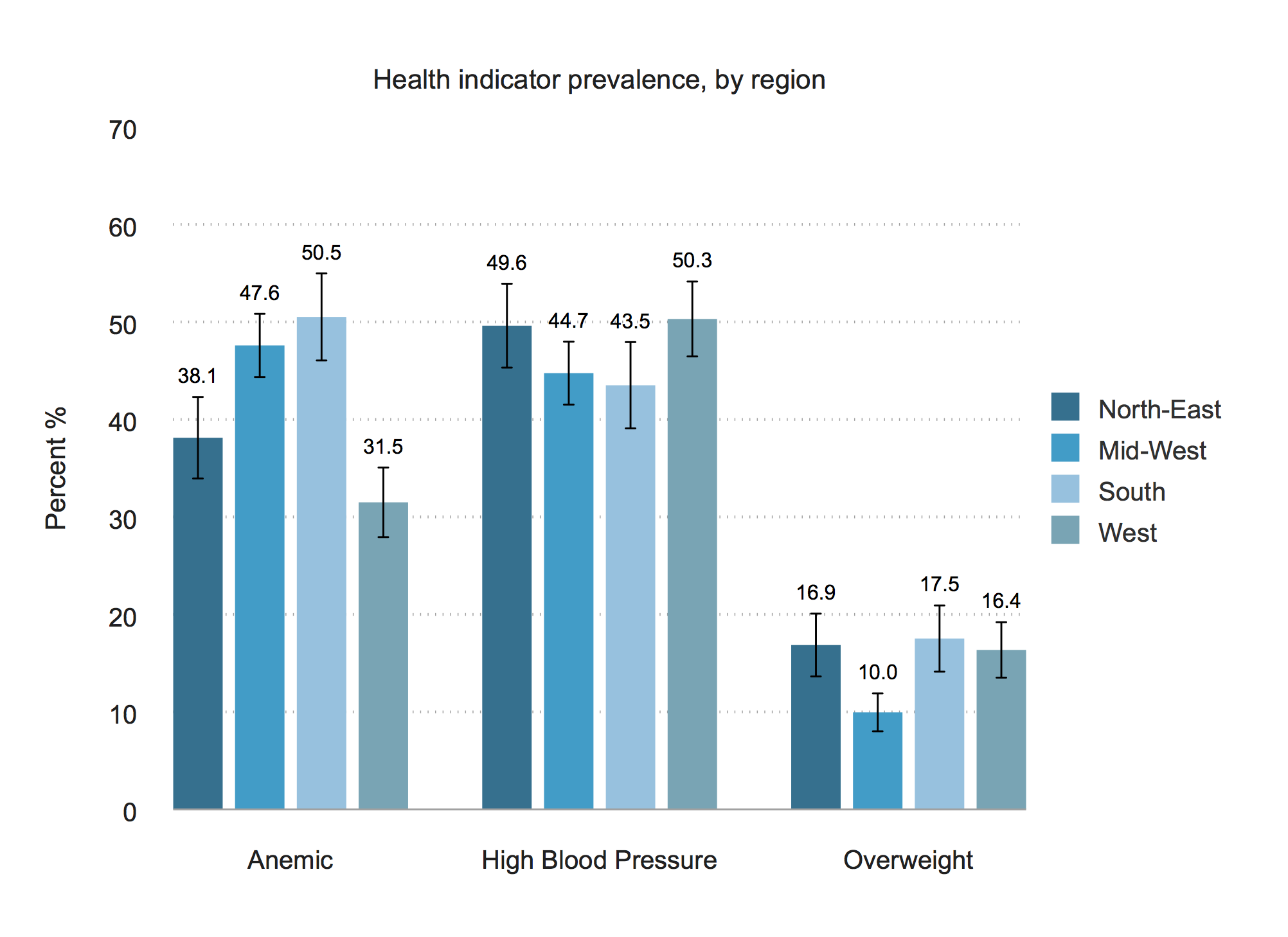
The chart
Stata
R
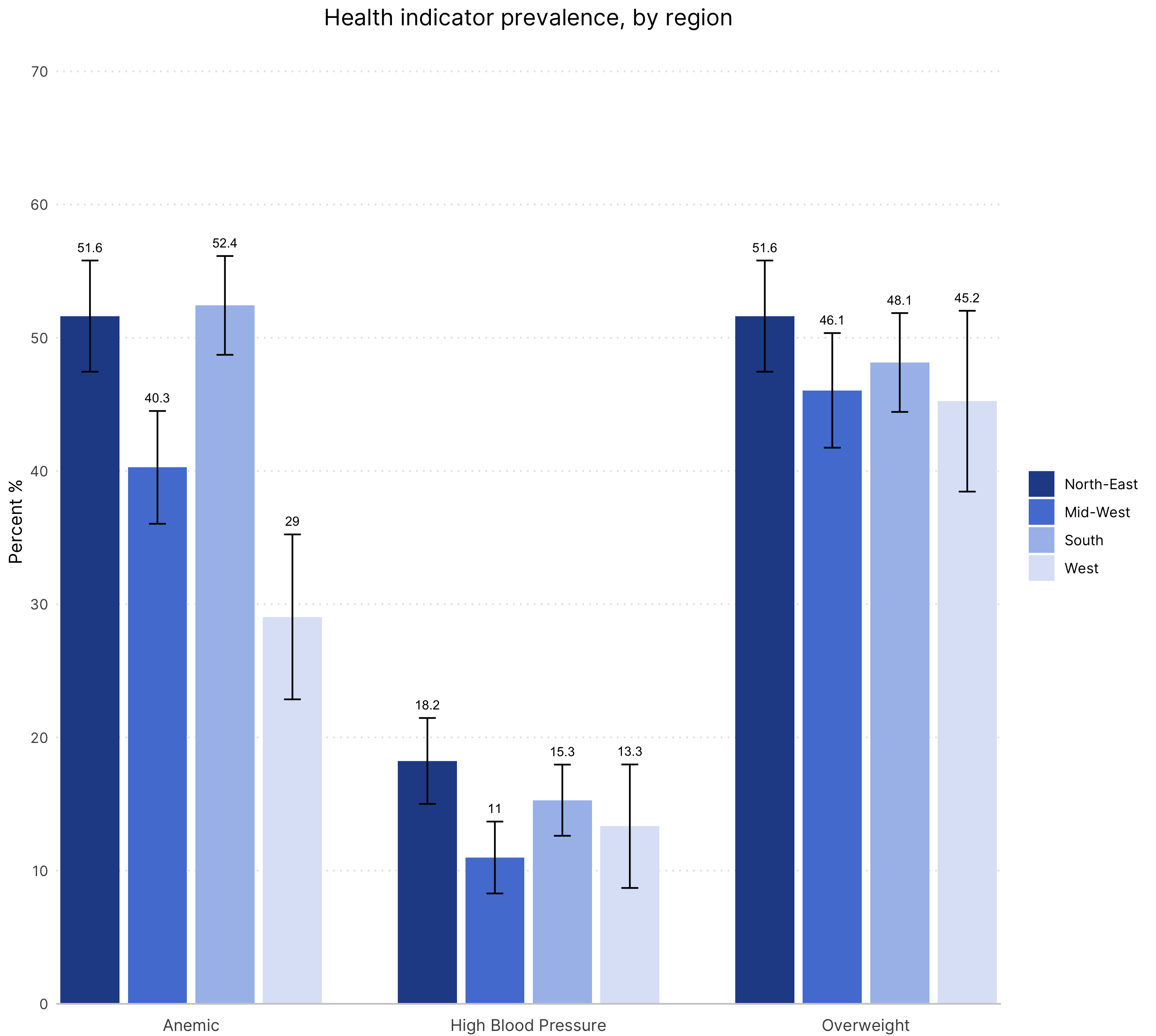
Data
Sample dataset in .dta format can be downloaded here. The R code uses the same dataset in .csv format, which can be found here.
The code
Stata
* Toy dataset
global data "/Users/crystalhuang/Dropbox (IDinsight)/Data Visualization Guide/3. Toy Data"
global output "/Users/crystalhuang/Dropbox (IDinsight)/Data Visualization Guide/2. Graph templates/graphs"
use "$data/toydata.dta", clear
* Set IDinsight scheme and font
set scheme idinsight
graph set window fontface "Arial"
* Graph 2 - BMI, Blood Pressure, Diabetes, by region
* Define locals for outcomes
local outcomes anemic overweight bp_high
* Get standard errors, upper and lower CIs for each outcome var
foreach y of local outcomes {
g se_`y'= .
g lo_`y'= .
g hi_`y'= .
* loop over grouping var
forval i= 1/4 {
mean `y' if region== `i'
mat A= r(table)
local se_`i'= A[2,1]
local lo_`i'= A[5,1]
local hi_`i'= A[6,1]
replace se_`y'= `se_`i'' if region== `i'
replace lo_`y'= `lo_`i'' if region== `i'
replace hi_`y'= `hi_`i'' if region== `i'
}
}
* Collapse to get height of each bar (outcome variable mean) by grouping var (region)
collapse (mean) `outcomes' (first) se_* lo_* hi_*, by(region)
g id= _n
* Rename so outcomes have the same prefix for reshaping long
foreach y of local outcomes {
ren `y' mean_`y'
}
* Reshape so it's one row per outcome, per group
reshape long mean se lo hi, i(id) j(outcome) string
replace outcome = subinstr(outcome, "_", "", 1)
* Set bar label format, make units into percent
foreach var in mean se hi lo {
replace `var'= `var'*100
}
format mean %4.1f
* Create x-axis spacing
gsort outcome +mean
g x= region if outcome== "anemic"
replace x= region + 5 if outcome== "overweight"
replace x= region + 10 if outcome== "bp_high"
* GRAPH: Vertical bars, grouped by region with CIs
#delimit;
twoway (bar mean x if region==1, barw(0.80))
(bar mean x if region==2, barw(0.80))
(bar mean x if region==3, barw(0.80))
(bar mean x if region==4, barw(0.80))
(rcap hi lo x, lwidth(thin) lcolor(black))
(scatter hi x, msym(none) mlab(mean) mlabpos(12) mlabgap(1) mlabsize(2.2) mlabcolor(black)),
ylabel(0(10)70, format(%4.0f))
xlabel(2.5 "Anemic" 7.5 "High Blood Pressure" 12.5 "Overweight", nogrid)
ytitle("Percent %")
xtitle("")
title("Health indicator prevalence, by region")
legend(region(lcolor(white)) order(1 "North-East" 2 "Mid-West" 3 "South" 4 "West") rows(4) pos(3))
yscale(lcolor(none)) ;
graph export "$output/bargraph_vertical.tif", replace ;
#delimit cr
R
# Vertical bar
################################# Initial Setup ################################
# Install required packages if they are not already in your system
packages <- c('tidyverse')
lapply(packages, function(i) {if(!i %in% installed.packages() == T)
{install.packages(i, dependencies = TRUE, repos='http://cran.rstudio.com/')}})
# Loading required packages
library("tidyverse")
# Setting working directory
setwd("~/Dropbox (IDinsight)/Data visualization library")
############################### Loading dataset ################################
mydata <- read_csv("Data/toydata.csv", show_col_types = FALSE)
################################ Data processing ###############################
# Changing the structure of the dataset from wide to long
mydata_long <- mydata %>%
select(personid, region, bp_high, anemic, overweight) %>%
pivot_longer(cols = c(3:5),
names_to = "disease",
values_to = "disease_present") %>%
arrange(personid)
# Converting the regions to factor variable so that we can maintain a specific
# legend order. The levels provided below dicates the order of the bars and
# the legend.
mydata_long$region <- factor(mydata_long$region,
levels = c("North-East", "Mid-West",
"South", "West"))
# Creating the means and CIs
# Assuming 95% CI
alpha = 0.05
# Creating means and CI and storing in my_sum tibble
my_sum <- mydata_long %>%
group_by(region, disease) %>%
summarise(n = n(),
mean = mean(disease_present, na.rm = T),
sd = sd(disease_present, na.rm = T)) %>%
mutate(se = sd/sqrt(n)) %>%
mutate(ic = se * qt((1-alpha)/2 + 0.5, n - 1)) %>%
arrange(disease)
############################### Creating the graph #############################
# Define axis and fill variable
x_values <- my_sum$disease
y_values <- my_sum$mean
fill_by <- my_sum$region
plot <- my_sum %>%
# Setting aesthetic which will be inherited by other geometric objects
ggplot(aes(x = x_values, y = y_values, fill = fill_by)) +
# "identity" selected for stat since we want the heights of the bars to
# represent values in the data. The default, "bin" makes the height of each
# bar equal to the number of cases in each group.
# position dodge applied so that the bars are not superimposed.
geom_bar(stat = "identity", position = position_dodge(width = 0.8), width = 0.7) +
# Setting the legend/scale colors
scale_fill_manual(values = c("#264D96", "#5480D6", "#A8BFEB", "#DEE5F7")) +
# Setting the x-axis labels. The expand option removes the space around the
# axis and the data. Please feel free to change the values and see how the
# plot is affected.
scale_x_discrete(labels = c("Anemic", "High Blood Pressure", "Overweight"),
expand = c(0, 0)) +
# Position dodge applied so that the error bars are not superimposed. The
# width can be tweaked to ensure that are positioned correctly on top of the
# bars
geom_errorbar(aes(ymin = y_values - ic, ymax = y_values + ic),
width = 0.20,
size = 0.5,
position = position_dodge(width = 0.8)) +
# Customizing the y-axis by changing the limits, axis tick values and labels
scale_y_continuous(expand = c(0, 0),
limits = c(0, max(y_values) + 0.2),
breaks = c(seq(from = 0, to = 0.7, by = 0.1)),
labels = c("0", "10", "20", "30", "40", "50", "60", "70")) +
# Adding the text which will display the mean of the bars. They are positioned
# 0.01 units on top of (mean + ic). This ensures that there is some space
# between the error bars and the text.
# As with previous geometric objects, position dodge has been applied here as
# well so that the mean is not superimposed.
geom_text(aes(y = y_values + ic + 0.01, label = round(y_values*100, 1)),
position = position_dodge(width = 0.8),
size = 2.8) +
# Graph title and axis labels
labs(title = "Health indicator prevalence, by region",
y = "Percent %")
############################# Formatting the graph #############################
plot +
theme_classic() +
# The following visual changes have been made:
# Removed x-axis title
# Made the x-axis line a light shade of gray
# Removed the y-axis line
# Created y-axis grid lines which are light gray and dotted
# Removed axis ticks
# Removed legend title
# Vertically aligned and resized x-axis text
# Center aligned and resized the plot title
theme(text = element_text(family = "Inter"),
axis.title.x = element_blank(),
axis.line.x = element_line(color = "grey80"),
axis.line.y = element_blank(),
panel.grid.major.y = element_line(colour = "grey90", linetype = "dotted"),
axis.ticks = element_blank(),
legend.title = element_blank(),
axis.text.x = element_text(vjust = -1, size = 10),
plot.title = element_text(hjust = 0.5, size = 14))
############################## Saving and exporting ############################
# Indicating the export folder and the image file name
export_folder <- "R/Bar graphs/Exports/"
img_name <- "bar_vertical_R_reviewed.png"
ggsave(paste(export_folder,img_name,sep = ""))
Other details
Stata
Credit: Crystal Huang
You must have IDinsight styles installed to get replicate the chart above.
R
Code written by Arkadeep Bandyopadhyay and reviewed by Sandra Alemayehu.
Colors for the graph have been selected from IDinsight’s brand guide.